GATE Bio-YODIE
Bio-YODIE is a named entity linking system derived from GATE YODIE. It links mentions in biomedical text to their referents in the UMLS, a large and popular medical terminology. Bio-YODIE has been developed as part of the KConnect project.
The screenshot below shows French Bio-YODIE having been run on a test document in the GATE GUI. The most important thing to note is that the selected annotations (in yellow) have been added to the document by the application, and features added to it, as shown in the popup window with reference to the mention "utilisateurs de drogues". The output annotations appear in the "bio" annotation set.
Many of the features relate to experimental approaches to disambiguation, and are in flux, but features of value to the end user include among others "inst" (the CUI) and "STY" (the semantic type, according to UMLS). CUI could for example be used to link to UMLS-based knowledge sources for semantic enrichment applications. STY might be used to find, for example, all disease names in the text.
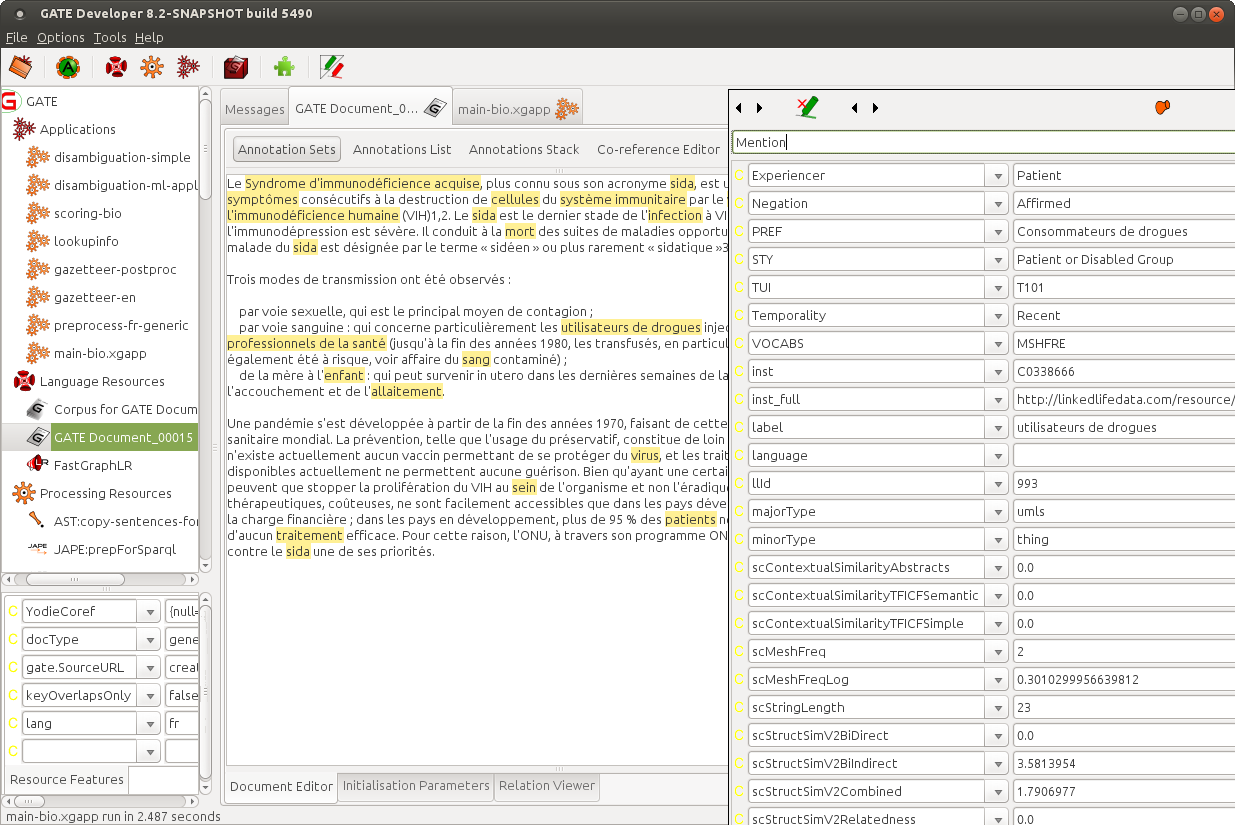
The application works by finding all mentions in the text that correspond to a label in UMLS. In some cases, more than one possible interpretation may be found for a piece of text. In that case, various knowledge sources are used to choose the best interpretation.
Bio-YODIE is also available as a service - you can try it out here. Please contact Xingyi Song (first initial dot surname at sheffield dot ac dot uk) for more information.
Bio-YODIE is also available open-source on GitHub.
To reference Bio-YODIE, please use Gorrell et al. 2018.
You can also find more information on our work using GATE for Biomedical text processing.





